What is 'Denoising' Anway?
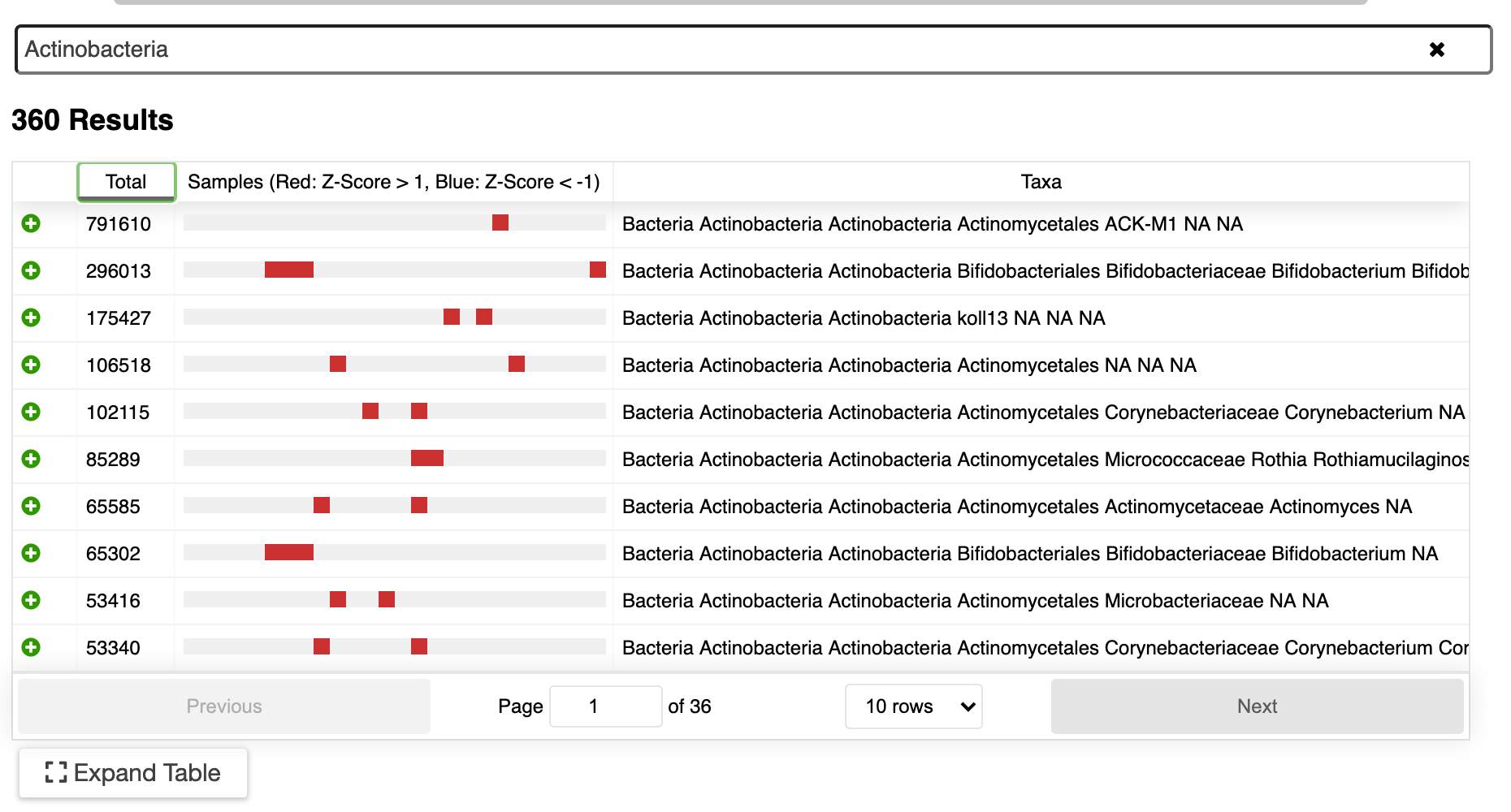
/Several different software packages such as DADA2, Deblur, and UNOISE use an error-correction technique that is applied to the sequenced amplicon (usually 16S, 18S, or ITS) reads. This ‘denoising’ step is used to increase correct taxonomic assignment of the sequenced amplicons.
Read More