Linear Discriminate (LEfSe) of Amplicon and Metagenome sequencing available in ERGO
/ERGO’s [Overbeek et. all 2003] rich suite of tools and workflows to examine amplicon and metagenome sequencing now includes Linear Discriminate Analysis using LEfSe [Segata et. al 2010]. In ERGO, LEfSe (Linear discriminate analysis Effect Size) enables researchers to discover biomarkers that most likely explain the differences between selected experimental groups.
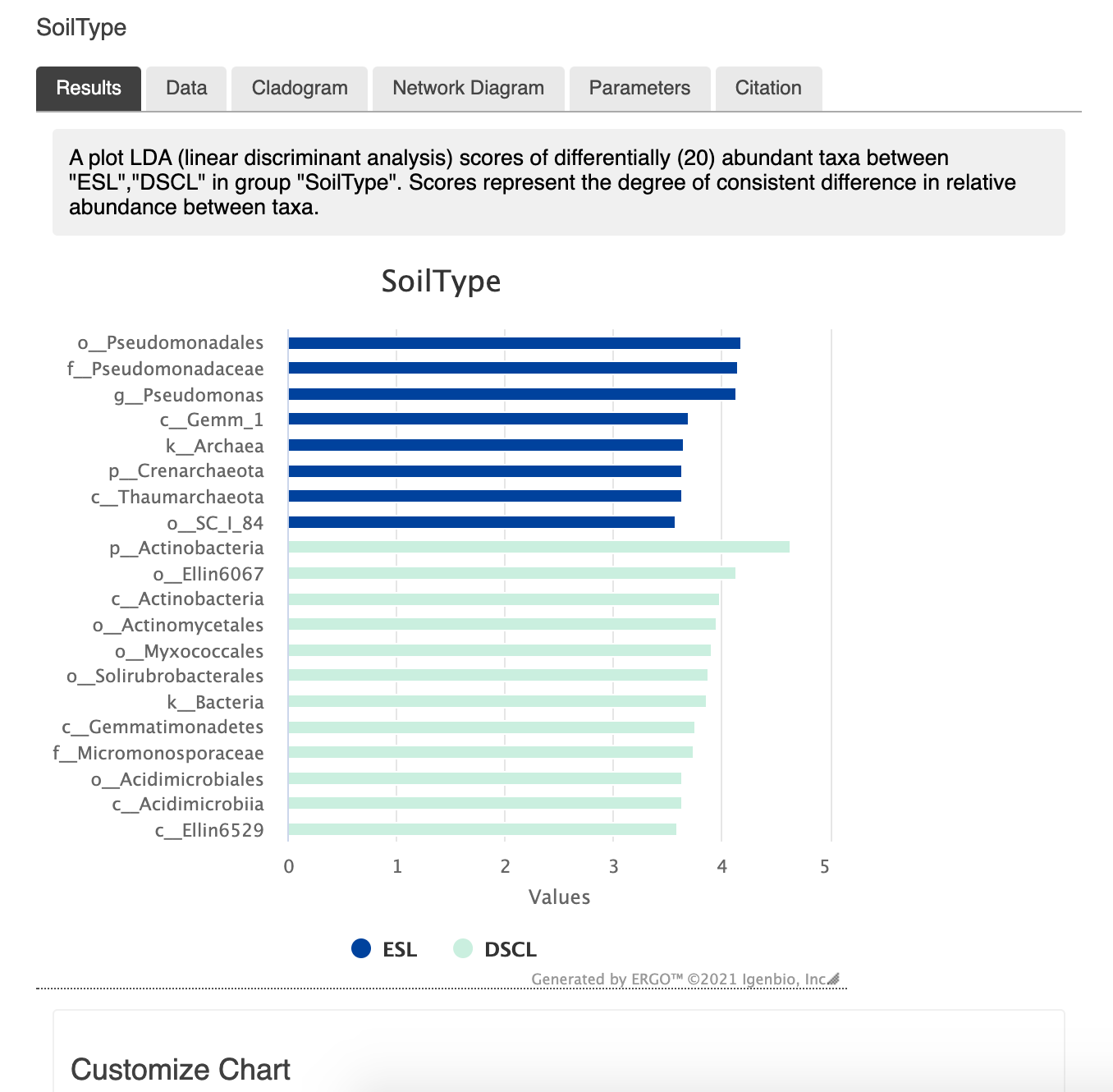
For example, from the plot below we can say that the genus Pseudomonas has a consistent differential abundance in ESL soil [Halter et. al 2020]
A bar plot of effect size in ERGO.
ERGO’s easy-to-use interface makes creating a LEfSe analysis straightforward - select your experimental condition and click “Create”. ERGO will automatically generate common plots such as bar plots of effect size and cladograms. All of ERGO’s plots offer many customization options so you can create the perfect publication-ready plot.
A cladogram plot generated by LEfSe and presented in ERGO.
Cladogram plots (example, above) summarize results simply. Each node colors and background shading are colored by the soil type (DSCL or ESL) that has the highest median. The size of the nodes are in proportion to that taxon’s abundance. This shows that the phylum Actinobacteria are significantly differentially abundant in DSCL soil and class Bacilli are significantly differentially abundant in ESL soil.
ERGO makes it simple to edit this plot - move the legends or increase the font size of the title.
A network diagram of taxonomic clades. Effect size is represent by the size of the nodes, while the color represents which condition the node is differentially abundant in.
Beyond the bar plots and cladograms, ERGO provides another interactive summarization of the LEfSe data - a Network Diagram (example above). Each node is a level in the taxonomic hierarchy. The color represents which class/group that node is differentially abundant in (ESL or DCSL in this example). The size of the node represents the effect size. The hierarchy is pruned if all further levels are classified to the same group, simplifying the representation.
This summarization enables us to quickly identify which taxa clades are statistically significant in each group. For example, phylum Actinobacteria immediately stands out as significant for DSCL, and class Gammaproteobacteria for ESL.
This diagram is fully interactive - change the node placement, size, and color. One can also filter the network to taxa of interest.
with ERGO’s LEfSe analysis, you can:
Create and manage multiple LEfSe Analysis
Easily create custom bar and column plots of effect size
Create and customize custom cladograms
Explore completely interactive network diagrams enabling easy discovery of metagenomic biomarkers
What to know more or give ERGO a try? Please use the form to contact us!
References
LEfSe
Nicola Segata, Jacques Izard, Levi Walron, Dirk Gevers, Larisa Miropolsky, Wendy Garrett, Curtis Huttenhower.
"Metagenomic Biomarker Discovery and Explanation"
Genome Biology, 2011 Jun 24;12(6):R60
ERGO
Overbeek R, Larsen N, Walunas T, D'Souza M, Pusch G, Selkov E Jr, Liolios K, Joukov V, Kaznadzey D, Anderson I, Bhattacharyya A, Burd H, Gardner W, Hanke P, Kapatral V, Mikhailova N, Vasieva O, Osterman A, Vonstein V, Fonstein M, Ivanova N, Kyrpides N. The ERGO genome analysis and discovery system. Nucleic Acids Res. 2003 Jan 1;31(1):164-71. doi: 10.1093/nar/gkg148. PMID: 12519973; PMCID: PMC165577.
DATA
Halter M, Vaisvil B, Kapatral V, Zahn J. Organic farming practices utilizing spent microbial biomass from an industrial fermentation facility promote transition to copiotrophic soil communities. J Ind Microbiol Biotechnol. 2020 Nov;47(11):1005-1018. doi: 10.1007/s10295-020-02318-z. Epub 2020 Oct 23. PMID: 33098066.