New ERGO Feature: Functional Prediction for 16S Amplicon Sequencing data
/We’ve added to ERGO’s rich suite of tools and workflows for 16S amplicon sequencing studies to include functional metagenome prediction utilizing Picrust2 [1].
Amplicon sequencing projects have many benefits, especially in turn-around-time and cost. One major weakness is that unlike whole shotgun metagenome sequencing, direct detection of gene functions isn’t possible. However, with ERGO’s Functional Prediction Workflow using Picrust2, the functional content of the community can be inferred.
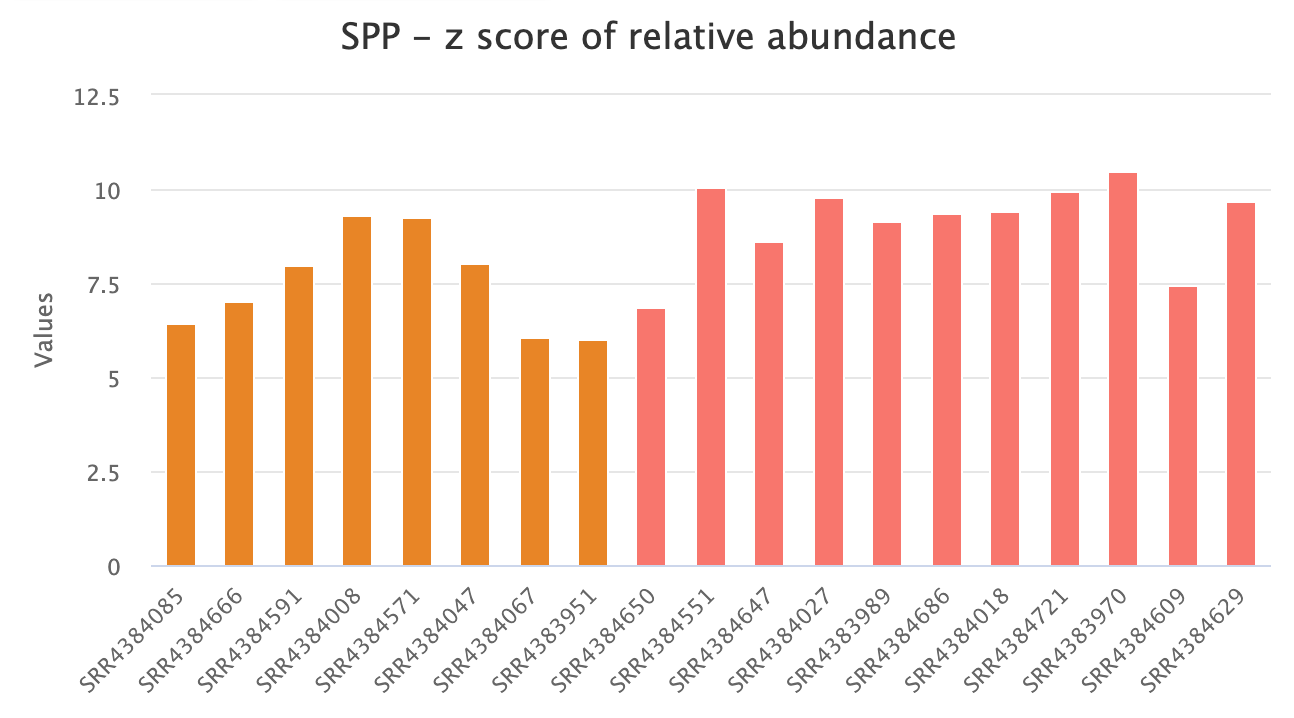
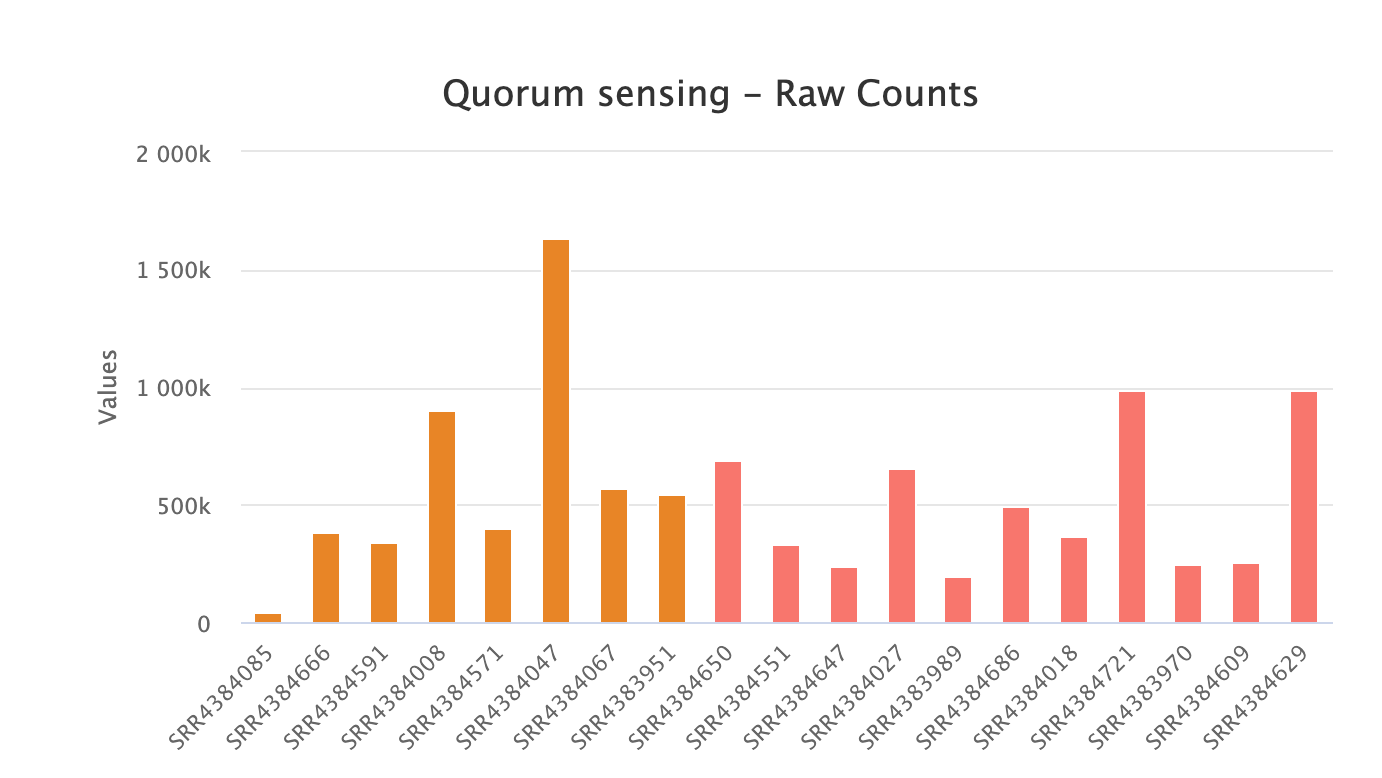
ERGO’s Workflow predicts, EC Numbers, KEGG Functions, and Pathways. Overall differences can be ascertained in a heatmap of metabolic categories, highlighting differences between sample groups. Each function can be individually explored revealing core differences between your samples.
Use the form below to start a free trial of ERGO.
Douglas, G.M., Maffei, V.J., Zaneveld, J.R. et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38, 685–688 (2020). https://doi.org/10.1038/s41587-020-0548-6


